|
|
PETER SZOLOVITS, Ph.D.; RAMESH S. PATIL, Ph.D.; and WILLIAM B. SCHWARTZ, M.D.; Cambridgeand Boston, Massachusetts
From the Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge,and Tufts University School of Medicine, Boston, Massachusetts In an attempt to overcome limitations inherent in conventional computer-aideddiagnosis, investigators have created programs that simulate expert human reasoning. Hopes that such a strategywould lead to clinically useful programs have not been fulfilled, but many of the problems impeding creation ofeffective artificial intelligence programs have been solved. Strategies have been developed to limit the numberof hypotheses that a program must consider and to incorporate pathophysiologic reasoning. The latter innovationpermits a program to analyze cases in which one disorder influences the presentation of another. Prototypes embodyingsuch reasoning can explain their conclusions in medical terms that can be reviewed by the user. Despite these advances,further major research and developmental efforts will be necessary before expert performance by the computer becomesa reality.
[MeSH terms: artificial intelligence; diagnosis, computer-assisted; expert systems; medical informatics;medical informatics applications; physicians; software design. Other indexing terms: disease models; hypotheses;pathophysiologic reasoning]
THE STEADY expansion of medical knowledge has made it more difficult forthe physician to remain abreast of medicine outside a narrow field. Consultation with a specialist is a solutionwhen the clinical problem lies beyond the physician's competence, but frequently expert opinion is either unavailableor not available in a timely fashion. Attempts have been made to develop computer programs that can serve as consultants(1-3). By the early 1970s it became clear that conventional tools such as flow charts, pattern matching, and Bayes'theorem were unable to deal with most complex clinical problems (4). Investigators then began to study the expertphysician to obtain detailed insights into the basic nature of clinical problem solving (5-8). The results derivedfrom such studies have subsequently formed the basis for computational models of the cognitive phenomena, and thesemodels have further been converted into so-called artificial intelligence programs (9-12).
Many of the early efforts to apply artificial intelligence methods to real problems, including medical reasoning,have primarily used rule-based systems (13). Such programs are typically easy to create, because their knowledgeis catalogued in the form of "if ... then..." rules used in chains of deduction to reach a conclusion.In many relatively well-constrained domains rule-based programs have begun to show skilled behavior (14). Thisis true in several narrow domains of medicine as well (14, 15), but most serious clinical problems are sobroad and complex that straightforward attempts to chain together larger sets of rules encounter major difficulties.Problems arise principally from the fact that rule-based programs do not embody a model of disease or clinicalreasoning. In the absence of such models, the addition of new rules leads to unanticipated interactions betweenrules and thus to serious degradation of program performance (16-18).
Given the difficulties encountered with rule-based systems, more recent efforts to use artificial intelligencein medicine have focused on programs organized around models of disease. Efforts to develop such programs haveled to substantial progress in our understanding of clinical expertise, in the translation of such expertise intocognitive models, and in the conversion of various models into promising experimental programs. Of equal importance,these programs have been steadily improved through the correction of flaws shown by confronting them with variousclinical problems.
We will focus on how improved representation of clinical knowledge and sophisticated problem-solving strategieshave advanced the field of artificial intelligence in medicine. Our purpose is to provide an overview of artificialintelligence in medicine to the physician who has had little contact with computer science. We will not concentrateon individual programs; rather, we will draw on the key insights of such programs to create a coherent pictureof artificial intelligence in medicine and the promising directions in which the field is moving. We will thereforedescribe the behavior not of a single existing program but the approach taken by one or another of the many programsto which we refer. It remains an important challenge to combine successfully the best characteristics of theseprograms to build effective computer-based medical expert systems. Several collections of papers (19-21) providedetailed descriptions of the programs on which our analysis is based.
A Basic Program for Clinical Problem-Solving
Any program designed to serve as a consultant to the physician must containcertain basic features. It must have a store of medical knowledge expressed as descriptions of possible diseases.Depending on the breadth of the clinical domain, the number of hypotheses in the database can range from a fewto many thousands. In the simplest conceivable representation of such knowledge, each disease hypothesis identifiesall of the features that can occur in the particular disorder. In addition, the program must be able to match whatis known about the patient with its store of information. Even the most sophisticated programstypically depend on this basic strategy.
The simplest version of such programs operates in the following fashion when presented with the chief complaintand when later given additional facts.
1. For each possible disease (diagnosis) determine whether the given findings are to be expected.
2. Score each disease (diagnosis) by counting the number of given findings that would have been expected.
3. Rank-order the possible diseases (diagnoses) according to their scores.
The power of such a simple program can be greatly enhanced through the use of a mechanism that poses questionsdesigned to elicit useful information. Take, for example, an expansion of the basic program by the following strategy:
4. Select the highest-ranking hypothesis and ask whether one of the features of that disease, not yet considered, is present or absent.
5. If inquiry has been made about all possible features of the highest-ranked hypothesis, ask about the features of the next best hypothesis.
6. If a new finding is offered, begin again with step 1; otherwise, print out the rank-ordered diagnoses and their respective supportive findings and stop.
Steps 1 through 3 contain a primitive evaluation of the available information, and steps 4 through 6 containan equally simple information-gathering strategy that determines what information to seek next. But such a programfails to capture many of the techniques responsible for expert performance. For example, the ranking process doesnot take into account how frequently particular features occur in a given disease. The program, furthermore, hasno knowledge of pathophysiology and is not able to take stock of the severity of an illness. The most serious problemis that each new finding sets into motion a search process tantamount to considering all disease states appearingin a textbook of medicine. Even for a high-speed computer this is not a practical diagnostic strategy and for thisreason research has turned to the study of how experts perform.
From Cognitive Models to Computer Programs
The physician's ability to sharply limit the number of hypotheses under active consideration atany one time is a key element in expert performance (5, 6, 9). Computer programs that use the strategies of expertscan accomplish this same goal and devote the bulk of their computational resources to the sophisticated evaluationof a small number of hypotheses.
Controlling the proliferation of hypotheses is only the first step in creating effective artificial intelligenceprograms. To deal with the circumstance in which one disease influences the clinical presentation of another, theprogram must also have the capacity to reason from cause to effect. Moreover, the required pathophysiologic knowledgemust be organized in a hierarchical fashion so that the information becomes more detailed as one progresses todeeper levels of the knowledge base. Quantitative information, or rough qualitative estimates, must also be addedto the causal links if the program is to separate the contribution of each of several disorders to a complex clinicalpicture.
The cognitive models that embody these principles provide the basis for computer programs that use the chiefcomplaint and other available information to reduce the range of diagnostic possibilities. The narrowing processcan be viewed as passive in that the program makes all possible progress without requesting further facts. Thepassive phase completed, the program moves to an active mode of posing questions to the physician. This processis interactive with each new fact stimulating additional analysis that further reduces the number of diagnosticpossibilities. In the following discussion, attention will be directed primarily to the passive narrowing processbecause this strategy plays a central role in clinical problem solving and because more is known about this processthan about the active collection of new information.
Passively Processing the Available Information
CONTROLLING THE NUMBER OF HYPOTHESES
One simple technique for limiting the number of active hypotheses consists of selecting from a large databaseonly those disorders for which there is evidence in the chief complaint. Limiting activation in this way is usefulbut rarely restricts the number of hypotheses to a small handful, typically three or four. An alternative and oftenmore effective strategy called triggering allows activation only in response to a finding highly suggestive ofa particular disease (9). For example, a history of vomiting blood will trigger "peptic ulcer" as anhypothesis; by contrast, the complaint of an occasional headache will not trigger "brain tumor." In thisscheme, findings other than triggers are used in the diagnostic process only when a particular hypothesis has alreadybeen activated. Unfortunately, even in this strategy a single trigger frequently generates an unmanageably largeset of hypotheses (22, 23). But, by using two findings, the behavior of the activation mechanism can often be improved.For example, the joint findings of hematuria and proteinuria can be used to activate a much narrower set of hypothesesthan will either finding alone. Adding more elements to the trigger will further restrict the number of hypothesesthat are activated, but the gain is sometimes achieved at a price; if a finding is improperly included in the triggeror a relevant finding is ignored, the possibility of a diagnostic error is considerably increased. Experimentalevidence suggests that a cluster of two or three findings provides the right balance between specificity and therisk of missing a diagnosis (24).
Facts obtained during the questioning phase may activate new hypotheses but frequently they also argue againstdiagnoses already under consideration. The new fact may be incompatible with a given hypothesis, such as a massiveamount of protein in the urine of a patient suspected of having uncomplicated chronic pyelonephritis, or it mayargue indirectly against a disease by strongly favoring a competing one. Under either circumstance, the hypothesiscan be removed from active consideration (9). Even a newly activated hypothesis can immediately be deactivatedif facts already available argue strongly against it.
Deactivation does not permanently exclude a hypothesis from consideration; the hypothesis may be reactivatedif additional supportive information is later obtained or if it must be explicitly ruled out in order to confirmsome other diagnosis (9).
AGGREGATES AND HIERARCHY IN NARROWING THE FOCUS
Even when the triggering process is combined with a mechanism for deactivation, it may not adequately controlthe proliferation of hypotheses. Under such circumstances, the diseases under consideration can be reduced in numberby grouping those of similar character (such as kidney diseases or infectious diseases) into a single hypothesisknown as an aggregate. Such a structure incorporates all of the findings that occur with particular frequency inthe cluster of diseases forming the aggregate. An aggregate cannot only stand in lieu of an unmanageably largenumber of diseases but can be organized into a hierarchy that facilitates analysis of the diagnostic problem. Thetop level aggregate of such a hierarchy contains all disorders under suspicion, and each lower level contains thesame disorders divided into successively smaller clusters. The program can then choose one of several strategiesto select the level within the hierarchy that provides the best focus for subsequent questioning.
Intermixed hierarchies: The first hierarchies used by artificial intelligence programs were intermixedin character (12, 25); each level in the hierarchy was organized around a different disease characteristic suchas duration of illness (acute or chronic), anatomical site, etiology, and so forth. In such a hierarchy, the programmust explore the sequence of characteristics in a predetermined fashion, typically from top to bottom or vice versa.But in many cases adherence to such a predetermined sequence will force the program into a grossly inefficientpattern of questioning and lead to poor diagnostic performance. Still another defect is that intermixed hierarchiescannot deal with multisystem diseases such as lupus erythematosus, scleroderma, or periarteritis nodosa (26).
Pure hierarchies: Because of these deficiencies, attention has shifted towards the use of so-called purehierarchies that incorporate only a single disease characteristic. A pure hierarchy for kidney diseases, for example,might be based on the anatomical site of involvement. Individual proximal and distal tubular diseases that appearat the lowest level of the hierarchy can be organized into an aggregate embodying all tubular diseases, and similaraggregates can be created for glomerular, interstitial, and vascular diseases. These aggregates can then be broughtto a higher level encompassing all kidney diseases. Such a structure can also be expanded to include nonrenal disorders.
Because a pure hierarchy has only a single organizing theme, the program can move across levels without difficultyand focus quickly on the level that merits further consideration. On the other hand, a diagnostic strategy basedon use of a single pure hierarchy is of no value when exploration of more than one clinical characteristic is required.This limitation has caused investigators to shift their attention to the use of multiple pure hierarchies. (27,28).
Reasoning with multiple pure hierarchies: Multiple pure hierarchies allow a program to explore a widerange of disease characteristics while preserving ease and clarity of analysis. Consider a patient who has ingesteda poison and is also oliguric. Multiple pure hierarchies allow the program to focus on those aspects of the patient'scondition most relevant to each significant initial fact, in this case identifying the cause of the illness andits pathophysiologic consequences as the prime issues, and then to integrate its understanding of the differentaspects of the case into an overall conclusion. First, the program takes all of the available facts and searchesthrough each hierarchy to identify the smallest set of hypotheses that it can validate; second, it searches acrossthe subsets drawn from each hierarchy to identify the diagnostic possibilities most worth pursuing.
Reasoning within an individual hierarchy can be accomplished by one of two means. The top-down Strategy is mostappropriate when little specific information is initially available, so that the most efficient approach consistsof moving from the general to the specific. The top-down strategy uses scoring methods to determine the goodness-of-fitbetween the observed manifestations and the highest-level disease hypothesis in a given hierarchy. If the hypothesisis found to be valid by some particular set of criteria, the program moves to the next level where there are twoor more aggregates, each encompassing a narrower range of diseases. If any one or several aggregates are foundto be valid, the entire process is repeated until a level is reached below which either validity cannot be shownor the total number of alternative hypotheses (usually four or five) becomes too large.
The bottom-up strategy is best used when the findings suggest a large number of specific diseases but do notprovide an organizing theme around which to formulate a differential diagnosis. The bottom-up strategy is initiatedby a triggering mechanism that selects the individual hypotheses that merit consideration. If these hypothesescannot be distinguished from one another on the basis of available information, the program moves to a higher levelin the hierarchy; this move is accomplished by replacing each group of individual diseases by the aggregate encompassingthem.
After having chosen the prime set of diagnostic possibilities within each hierarchy, the program moves intothe second phase in which it looks across the subsets to identify those diseases on which further questioning shouldfocus. These diseases are found by identifying those disorders that appear in two or more subsets (27). For example,in the oliguric patient who is known to have ingested a poison, the intersection between the prime disease setsin the anatomic and etiologic hierarchies will yield a tentative diagnosis of acute renal failure of nephrotoxicorigin. In more complex cases, several diseases will emerge from this process. The computation of such an intersection,although seemingly simple, is a fairly complex programming task. Skilled physicians, on the other hand, carry outthis process rather easily, probably because they have previously explored so many search paths that they knowin advance the answers. A similar pre-exploration has recently been exploited to good effect in programs that makeuse of several hierarchies (27).
Dealing with Multiple Disorders The strategies thus far considered assume that the patient has one disease. If several disorders are present,the problem is more complex. Additional difficulties arise if the several possible diseases have findings in commonor if one disorder influences the presentation of another. The challenge posed by several disorders pushes existingartificial intelligence programs to their conceptual and computational limits.
DISORDERS THAT DO NOT INFLUENCE EACH OTHER’S CLINICAL PRESENTATION
Nearly all early programs that dealt with several disorders were successful in diagnosing only diseases withoutoverlapping findings. These programs assumed that all hypotheses were competitors and attempted to identify thesingle most likely diagnosis (22). Only after the first diagnosis was confirmed did they attempt to make a seconddiagnosis based on the residual findings, a process that was repeated as long as there were findings not accountedfor by an already confirmed diagnosis. Such a sequential approach contains a major flaw: because the program initiallyhas no way of recognizing that more than one disorder exists, findings that are not relevant to the primary disordercan easily confound the diagnostic process. For example, in a patient with both chronic glomerulonephritis andan acute myocardial infarction, the program will try to attribute all clinical manifestations to each disease.It may, therefore, dismiss the diagnosis of chronic glomerulonephritis simply because it cannot account for severechest pain.
A partial solution to this problem can be achieved if one assumes that coexisting disorders should, in general,account for a larger set of observed findings than either alone. The Internist-l program (12) exploits this idea.First, all active hypotheses are rank-ordered and the leading hypothesis is taken as the focus of the diagnosticprocess; any diseases that account for findings not already explained by the leading hypothesis are removed fromthe active list and put aside for later consideration. The hypotheses remaining on the active list are consideredcompetitors of both the leading hypothesis and each other. The program then pursues various standard strategiesfor information gathering to arrive at a diagnosis. It then subsequently turns to the disorders that have beenset aside earlier and carries out the same process of differential diagnosis.
This ability to partition the sets of diseases and findings is the key to Internist- l's ability to diagnosecorrectly many of the cases drawn from clinico-pathologic conferences (12, 27). But even such a partitioning algorithm
cannot deal with two diseases whose findings overlap appreciably. If all observed findings are common to bothdiseases, the program will incorrectly consider the two to be competitors. Thus after confirming the presence ofone disease, it will ignore the other because all shared findings have been accounted for. Moreover, the programcannot deal with one disorder that has altered the clinical presentation of another (29). Consider a patient withacute renal failure of some days' duration whose illness is complicated by severe vomiting. If the serum potassiumconcentration was normal or low and the program expected an elevated serum potassium level, the program would notbe able to make the correct diagnosis.
DISORDERS WHOSE FINDINGS OVERLAP OR INTERACT
To deal with diseases whose findings overlap or interact, a program's best strategy is to use pathophysiologicreasoning that links diseases and findings through a network of causal relations. Through this mechanism, whichemulates expert human performance, the program can create a composite hypothesis that attempts to explain all ofthe clinical findings. If several combinations of diseases are consistent with available information, several competingcomposite hypotheses must be constructed. This process cannot be done in the same fashion as with individual diseasehypotheses. Descriptions of individual diseases can be created in advance and made available on demand. Potentialcomposite hypotheses, because they are extremely large in number, must instead be fashioned on an individual basisfrom the findings in a particular case.
The core of a composite hypothesis for a given patient is constructed by bringing together the set of abnormalstates (such as pulmonary insufficiency, hypertension, acidosis) that make up the overall clinical picture (28).To this core are added its possible underlying causes and the mechanisms that bring about its clinical manifestations.A representative composite hypothesis is shown in level I of Figure 1, which shows the simplest causal networkaccounting for the acidosis and hypokalemia induced by a combination of severe diarrhea and a moderate degree ofvomiting. Each possible explanation for the electrolyte disorders, such as renal failure or diabetic ketoacidosis,is represented in the program as a competing composite hypothesis. If no single cause adequately accounts for theseverity of all the findings, the program will conclude that more than one cause must be present. The program thenuses pathophysiologic reasoning to estimate the effects of interactions among the possible causes. Interactionsamong diseases can be estimated more precisely by supplementing the causal links with quantitative informationdescribing the magnitude of each cause and effect (29, 30).
Even rough qualitative estimates (such as slight, moderate, or severe) can assist in determining whether a singlediagnosis is consistent with known findings (31, 32). If, for example, a patient with mild congestive heart failureis found to have massive edema, the program will suspect that a second disorder (such as the nephrotic syndrome)is present. If additional evidence supporting a second cause can be found, it will be added to the composite hypothesis.If no such explanation is forthcoming, the program will consider laboratory error or a faulty patient history.
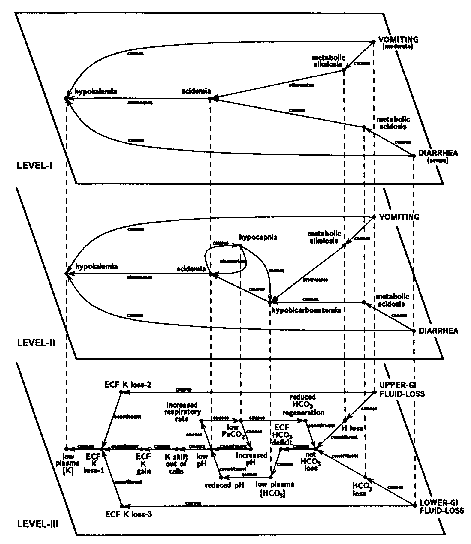
Figure 1. Three levels of detail in a composite hypothesis describing a patient with bothacid-base and potassium abnormalities induced by diarrhea and vomiting. The circles represent clinical and pathophysiologicstates and solid lines show the relationships among them. Relationships are labeled to indicate that one stateeither causes or attenuates another, or that two states are c6nstituents of a third. Etiologies are in capitalletters. and the dashed vertical lines show corresponding states at the different levels of detail. Each node inthe figure is associated with numerical values (not shown) reflecting the severity and duration of the particularstate. The clinical associations shown on level I are more fully elaborated by level II. which depicts the homeostaticadjustments in a patient with diarrhea and vomiting. Level III provides an even more detailed description of howthe organism responds to gastrointestinal losses (28, 29). GI = gastrointestinal; ECF = extracellular fluid.
To differentiate among the possible causes, all current information about the patient is used to generate a differentpossible series of events (scenarios) that might have led to the current clinical picture. Each scenario predictsvarious findings, some perhaps not yet found, that would be expected if a given disease were present (11, 30).The findings in each scenario are then compared so that differential diagnostic features can be identified andquestioning focused on them. For example, urinary sodium concentration will be identified as a feature that canhelp distinguish between oliguria due to acute tubular necrosis and that due to dehydration and volume depletion.
HIERARCHICAL ORGANIZATION IN CAUSAL REASONING
The more detailed the causal reasoning, the greater the price in terms of computational costs. Such costs canbe minimized, however, by organizing knowledge into layers of increasing detail. A system based on such a knowledgebase can select the most appropriate level at which to operate, using efficient, shallow reasoning in simple casesand resorting to expensive, detailed reasoning only when there is no alternative (28). A small hierarchical compositehypothesis, showing three levels of detail, is shown in Figure 1 (28). The shallow reasoning of level I, describedearlier, is more fully elaborated by level II, which shows the homeostatic adjustments in a patient with diarrheaand vomiting. Level III provides a more detailed description of how the organism responds to gastrointestinal losses.
Many of the ideas discussed thus far have been tested in experimental programs, but no program has yet succeededin integrating the various mechanisms required to produce a useful and reliable expert consultant.
Information Gathering and Reaching Diagnostic Conclusions Once the passive component of the program has reduced the number of hypotheses as much as possible, the activemode of questioning begins. The diagnostic strategies confirm, eliminate, and differentiate are derived, as inthe first portion of the program, from analysis of expert performance (5, 6, 12). The choice of a particular strategyis based on the following criteria. If a single hypothesis is the leading candidate by a wide margin, the programwill gather data designed to confirm the diagnosis or at least to give it further credence. If no such data canbe obtained readily and safely, the program will try to elicit information that can eliminate one or more of thecompeting diagnoses. Differentiation, the last of the three strategies, is generally used when only two hypothesesare under active consideration; the purpose is to gather information that favors one diagnosis while arguing againstthe other.
In many clinical situations, however, an optimal strategy cannot be chosen using the simple criteria just described(30, 33); instead, one must develop a plan for questioning based on possible answers to the series of questionsthat might be posed. One recent approach consists of developing a coherent plan for information gathering basedon stored knowledge of diagnostic strategies (30, 33, 34). For example, when the program is trying to differentiatebetween renal and essential hypertension, it would note that the diagnosis of essential hypertension is typicallymade by exclusion. On this basis, the program will develop a strategy designed to confirm the diagnosis of renalhypertension rather than differentiate between the two disorders. To accomplish this goal, the program will establishvarious sequences of possible questions and answers and then choose the line of questioning that looks most promising(4, 30).
REACHING A DIAGNOSTIC CONCLUSION
A pathognomonic abnormality provides the easiest path to diagnosis, but such findings are extremely uncommon.Moreover, even such a finding must be viewed with caution because of the possibility of error; corroboration ofa pathognomonic finding by other data is necessary before a conclusion can be reached.
If the questioning process has been completed and the diagnosis is still in doubt, the program rank-orders theset of hypotheses still under active consideration and reports the results to the user. Several numerical scoringschemes have been used in such a scoring process but none have proved completely satisfactory. The commonest schemequantifies the frequency with which each finding is associated with a given disease (9, 12, 22, 27) and simplysums the weights assigned to such findings. A more sophisticated version of this strategy makes formal use of Bayes'theorem (35-37). The diagnostic investigation is typically terminated when the score, or a value for the probability,has reached some predetermined threshold (9, 12, 35, 38). Available evidence indicates that humans have great difficultyin making reliable probabilistic judgments and calculations (39), suggesting that skilled physicians reach diagnosticclosure by unidentified strategies.
A program may not reach a diagnostic threshold even after gathering all the useful information that can be obtainedwithout using studies that impose risk or pain. At this point, decision analysis can be used to decide whethersuch studies should be done or whether treatment should be initiated even in the face of considerable uncertainty(40). The response to treatment will, of course, sometimes provide the best means of arriving at a firm diagnosis.
Discussion Most approaches to computer-assisted diagnosis have, until the past few years, been based on one of three strategies-flowcharts (1, 2, 41), statistical pattern-matching (42), or probability theory (4, 35, 43, 44). All three techniqueshave been successfully applied to narrow medical domains, but each has serious drawbacks when applied to broadareas of clinical medicine. Flow charts quickly become unmanageably large. Further, they are unable to deal withuncertainty, a key element in most serious diagnostic problems. Probabilistic methods and statistical pattern-matchingtypically incorporate unwarranted assumptions, such as that the set of diseases under consideration is exhaustive,that the diseases under suspicion are mutually exclusive, or that each clinical finding occurs independently ofall others (22). In theory, these problems could be avoided by establishing a database of probabilities that copeswith all possible interactions (37). But gathering and maintaining such a massive database would be a nearly impossibletask. Moreover, all programs that rely solely on statistical techniques ignore causality of disease and thus cannotexplain to the physician their reasoning processes nor how they reach their diagnostic conclusions.
Programs using artificial intelligence techniques have several major advantages over programs using more traditionalmethods. These programs have a greater capacity to quickly narrow the number of diagnostic possibilities, theycan effectively use pathophysiologic reasoning, and they can create models of a specific patient's illness. Suchmodels can even capture the complexities created by several disease states that interact and overlap. These programscan also explain in a straightforward manner how particular conclusions have been reached (33, 45). This latterability promises to be of critical importance when expert systems become available for day-to-day use; unless physicianscan assess the validity of a program's conclusions, they cannot rely on the computer as a consultant. Indeed, arecent survey has shown that a program's ability to explain its reasoning is considered by clinicians to be moreimportant than its ability to arrive consistently at the correct diagnosis (46). An explanatory capability willalso be required by those responsible for correcting errors or modifying programs; as programs become larger andmore complicated, no one will be able to penetrate their complexity without help from the programs themselves.
Causal, quantitative reasoning also leads to programs that can plan and manage therapy. Past events can be usednot only to predict current findings but to anticipate the possible future evolution of an illness and the consequencesof particular therapeutic actions (47, 48). Such capabilities provide the framework for expanding computer programsbeyond their conventional bounds as diagnostic aids.
Progress toward developing practical consulting programs has been slow despite the rapid increase in our understandingof how experts solve problems. Experience shows that 5 years is required to incorporate a new cognitive model intoan artificial intelligence program and to test it adequately. Two major factors have prevented more rapid implementation.First, a large amount of detailed medical knowledge must be gathered even when one is dealing with a relativelynarrow clinical domain. Second, newer cognitive models are so complex that their implementation typically posesa major technical challenge.
Even if the various problems in implementation can be solved, further obstacles will impede the developmentof programs that are ready for routine clinical use. Decisions must be made concerning acceptable performance levels(1) and extensive debugging and in-hospital testing must be done to assure that the standards are being met.
Fortunately, even before the advent of fully functional computer programs that can act as sophisticated consultantson the most difficult medical problems, the fruits of artificial intelligence research can be applied in less taxingmedical settings. Two recent programs, for example, combine the scoring methods of Internist-l (12) and databasesthat link diseases with their manifestations to generate lists of hypotheses that may be worthy of detailed consideration(49, 50). Other artificial intelligence programs applied in narrow medical domains have also proved to have practicalvalue, in applications ranging from laboratory data interpretation to protocol-based patient management (51-53).Although only a few such programs are currently available, the evidence suggests that the continued developmentof artificial intelligence techniques will eventually give the computer a major role as an expert consultant tothe physician.
ACKNOWLEDGMENTS: The authors thank William Bennett, M.D., Harvard Medical School, and NicolaosMadias. M.D.. New England Medical Center Hospitals, for their insightful comments and criticisms.
Grant support: in part by National Institutes of Health grants R24 RR 01320 from the Divisionof Research Resources, ROI HL 33041 from the National Heart, Lung and Blood Institute, RO1 LM 04493 from the NationalLibrary of Medicine; and from the Robert Wood Johnson Foundation and the Commonwealth Fund. The views expressedare those of the authors and do not necessarily represent the views of any of the granting agencies.
Requests for reprints should be addressed to Peter Szolovits. Ph.D.; MIT Laboratory forComputer Science, 545 Technology Square; Cambridge, MA 02139.
References
1. SCHWARTZ WB. Medicine and the computer: the promise and problems of change. N Engl J Med. 1970;283:1257-64.
2. LUSTED LB. Introduction to Medical Decision Making. Springfield, Illinois: Thomas; 1968.
3. JACQUEZ JA, ed. Computer Diagnosis and Diagnostic Methods. Springfield; Illinois: Thomas; 1972.
4. GORRY GA. Computer-assisted clinical decision making. Methods Jnf Med. 1973;12:45-51.
5. KASSIRER JP, GORRY GA. Clinical problem solving: a behavioral analysis. Ann Intern Med. 1978;89:245-55.
6. ELSTEIN AS, SHULMAN LS, SPRAFRA SA. Medical Problem Solving: An Analysis of Clinical Reasoning. Cambridge: Harvard University Press; 1978.
7. SWANSON DB, FELTOVICH PJ, JOHNSON PE. Psychological analysis of physician expertise: implications for design of decision support systems. In: SHIRES DB, WOLF H, eds. Proceedings of the Second World Conference on Medical Informatics. New York: North Holland; 1977:161-4.
8. KUIPERS BJ, KASSIRER JP. Causal reasoning in medicine: analysis of protocol. Cognitive Sci 1984;8:363-85.
9. PAUKER SG. GORRY GA, KASSIRER JP, SCHWARTZ WB. Towards the simulation of clinical cognition: taking a present illness by computer. Am J Med. 1976;60:981-96.
10. SHORTLIFFE. EH. Computer-Based Medical Consultations: MYCIN. New York: Elsevier; 1976.
11. WEISS SM, KULIKOWSKI CA, AMAREL S, SAFIR A. A model-based method for computer-aided medical decision making. Artif Intell.1978;11:145-72.
12. MILLER RA, POPLE HE JR, MYERS JD. Internist-I, an experimental computer-based diagnostic consultant for general internal medicine. N EngI J Med. 1982;307:468-76.
13. DUDA RO, SHORTLIFFE EH. Expert systems research. Science.1983;220:261-8.
14. BUCHANAN BG. Expert systems: working systems and the research literature. Expert Systems. 1986;3:32-51.
15. BUCHANAN BG, SHORTLIFFE EH, eds. Rule-Based Expert Systems: The MYCIN Experiments of the Stanford Heuristic Programming Project. Reading: Addison-Wesley Publishing Co.; 1984.
16. CLANCEY WJ, LETSINGER R. Neomycin: reconfiguring a rule-based expert system. In: Proceedings of the Seventh International Conference on Artificial Intelligence. Los Altos, California: M. Kaufmann Publishers; 1981:829-36.
17. DAVIS R. Expert systems: Where are we? and where do we go from here? AI Magazine. 1982;3:3-22.
18. SCHWARTZ WB, PATIL RS, SZOLOVITS P. Artificial intelligence in medicine: where do we stand? N Engl J Med. 1987;316:685-8.
19. SZOLOVITS P, ed. Artificial Intelligence in Medicine. Boulder: Westview Press; 1982;51.
20. CLANCEY WJ, SHORTLIFFE EH. eds. Readings in Medical Artificial Intelligence: The First Decade. Reading, Massachusetts: Addison-Wesley; 1984.
21. REOGIA JA, TUHRIM S. Computer-Assisted Medical Decision Making. New York: Springer-Verlag; 1985.
22. SZOLOVITS P, PAUKER SG. Categorical and probabilistic reasoning in medical diagnosis. Artif lntell 1 978;1l:115-44.
23. PAUKER SG, SZOLOVITS P. Analyzing and simulating taking the history of the present illness: context formation. In: SCHNEIDER W, SAGALL-HEIN AL, eds. Computational Linguistics in Medicine. Amsterdam: North Holland; 1977:109-18.
24. SHERMAN HB. A Comparative Study of Computer-Aided Clinical Diagnosis of Birth Defects. (Technical Rep. TR-283). Cambridge: MIT Laboratory for Computer Science; 1981:83-5,114,126-8.
25. CHANDRASEKARAN B, MITTAL S. Conceptual representation of medical knowledge for diagnosis by computer: MDX and related systems. In: YOVITS MC, ed. Advances in Computers. New York: Academic Press; 1983:217-93.
26. POPLE HE JR. The formation of composite hypotheses in diagnostic problem solving: an exercise in synthetic reasoning. In: Proceedings of the Fifth International Joint Conference on Artificial Intelligence. Los Altos, California: M. Kaufmann: 1977:1030-7.
27. POPLE HE JR. Heuristic methods for imposing structure on ill-structured problems: the structuring of medical diagnostics. In: SZOLOVITS P, ed. Artificial Intelligence in Medicine. Boulder: Westview Press; 1982:119-90.
28. PATIL RS. Causal Representation of Patient Illness for Electrolyte and Acid-Base Diagnosis. (Technical Rep. TR-267.) Cambridge: MIT Laboratory for Computer Science; 1981.
29. PATIL RS, SZOLOVITS P, SCHWARTZ WB. Causal understanding of patient illness in medical diagnosis. In: Proceedings of the Seventh International Joint Conference on Artificial Intelligence. Los Altos, California: M. Kaufmann; 1981:893-9.
30. PATIL RS, SZOLOVITS P, SCHWARTZ WB. Information acquisition in diagnosis. In: Proceedings of the National Conference on Artificial Intelligence. Los Altos. California: M. Kaufmann; 1982:345-8.
31. KUIPERS RI. Qualitative Simulation in Medical Physiology: A Progress Report. (Technical Rep. TM-280.) Cambridge: MIT Laboratory for Computer Science; 1985.
32. BORROW DG, ed. Qualitative Reasoning about Physical Systems. Amsterdam: North Holland; 1984.
33. CLANCEY WJ. The epistemology of a rule-based expert system: a framework for explanation. Artif Intell. 1983;20:215-51.
34. CHANDRASEKARAN B. Generic tasks in knowledge-based reasoning: high-level building blocks for expert system design. IEEE Expert. 1986;1:23-30.
35. GORRY GA, KASSIRER JP, ESSIG A, SCHWARTZ WB. Decision analysis as the basis for computer-aided management of acute renal failure. Am J Med. 1973;55:473-84.
36. COOPER GF. A diagnostic method that uses causal knowledge and linear programming in the application of Bayes' formula. Comput Methods Programs Biomed. 1986;22:223-37.
37. PEARL J. Fusion, propagation, and structuring in belief networks. Artif Intell. 1986;29:241-88.
38. PAUKER SG, KASSIRER JP. The threshold approach to clinical decision making. N Engl J Med. 1980;302:1109-17.
39. TVERSKY A, KAHNEMAN D. Judgment under uncertainty: heuristics and biases. Science. l974;185:l124-31.
40. PAUKER SG, KASSIRER JP. Therapeutic decision making: a cost benefit analysis. N Engl J Med. 1975;293.229-34.
41. BLEICH HL. Computer-based consultation: electrolyte and acid-base disorders. Am J Med. 1972;53:285-91.
42. ROSATI RA, MCNEER JF, STARMER CF, MITTLER BS, MORRIS JJ JR, WALLACE AG. A new information system for medical practice. Arch Intern Med. 1975;135:1017-24.
43. DE DOMBAL FT, LEAPER DJ, STANILAND JR, MCCANN AP, HORROCKS JC. Computer-aided diagnosis of abdominal pain. Br Med J 1972;2:9-l3.
44. WEINSTEIN MC, FINEBERG HV. Clinical Decision Analysis. Philadelphia: W. B. Saunders Co.; 1980.
45. SWARTOUT WR. XPLAIN: a system for creating and explaining expert consulting programs. Artif lntell. 1983;21:285-325.
46. TEACH RL, SHORTLIFFE EH. An analysis of physicians' attitudes. In: BUCHANAN BG, SHORTLIFFE EH, eds. Rule-Based Expert Systems: The MYCIN Experiments of the Stanford Heuristic Programming Project. Reading, Massachusetts: Addison-Wesley; 1984:635-52.
47. LONG WJ. Reasoning about state from causation and time in a medical domain. In: Proceedings of the National Conference on Artificial Intelligence. Los Altos, California: M. Kaufmann; 1983:251-4.
48. LONG WJ, NAIMI S, CRISCITIELLO MG, PAUKER SG, SZOLOVITS P. An aid to physiological reasoning in the management of cardiovascular disease. In: RIPLEY KL, ed. Proceedings of the 1984 Computers in Cardiology Conference. Los Angeles: IEEE Computer Society Press; 1984:3-6.
49. MILLER RA, MCNEIL MA, CHALLINOR SM, MASARIE FE JR, MYERS JD. The Internist-I/Quick Medical Reference project-status report. West J Med. 1986;145:816-22.
50. BARNETT GO, CIMINO JJ, HUPP JA, HOFFER EP. DXplain: an evolving diagnostic decision-support system. JAMA. 1987;258:67-74.
51. WEISS SM, KULIKOWSRI CA, GALEN RS. Developing microprocessor-based expert models for instrument interpretation. In: Proceedings of the Seventh International Joint Conference on Artificial Intelligence. Los Altos, California: M. Kaufmann; 1981:853-5.
52. AIKINS JS, KUNZ JC, SHORTLIFFE EH, FALLAT RJ. PUFF: an expert system for interpretation of pulmonary function data. Comput Biomed Res. 1983;16:199-208.
53. HICKAM DH, SHORTLIFFE EH, BISCHOFF MB, SCOTT AC, JACOBS CD. The treatment advice of a computer-based cancer chemotherapy protocol advisor. Ann Intern Med. 1985;103:928-36.
from ANNALS OF INTERNAL MEDICINE Vol.108; No.1, pages 80-87. January 1988.
来自圈子: 东游西逛 |
|